Overview of Interactome3D
Video tutorials
|
|
|
| Overview | Submit your data and check the results | Browse Interactome3D pre-built datasets |
|---|---|---|
|
|
|
| Query with a list of proteins | Download Interactome3D data |
Step-by-step tutorials
- Submit your data and check the results
- Browse Interactome3D pre-built datasets
- Query Interactome3D with a list of proteins
- Download Interactome3D data
Tutorial: Submit your interactions [top]
1 |
Interactome3D allows you to retrieve structures and build
homology models for a set of interactions you provide. Go to http://interactome3d.irbbarcelona.org and paste your interactions into the left textbox or upload them as a file (an example is available at human_interactions_example.txt). Your interactions should be provided as a list of pairs of Uniprot accession codes (AC). |


|
2 |
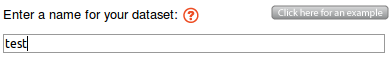
You have to choose a name for your dataset and introduce your email address. |
|
3 |
Once you have entered this data, click on the submit button. The Interactome3D pipeline will start processing your data and send you an email as soon as the results are available. |
|
4 |
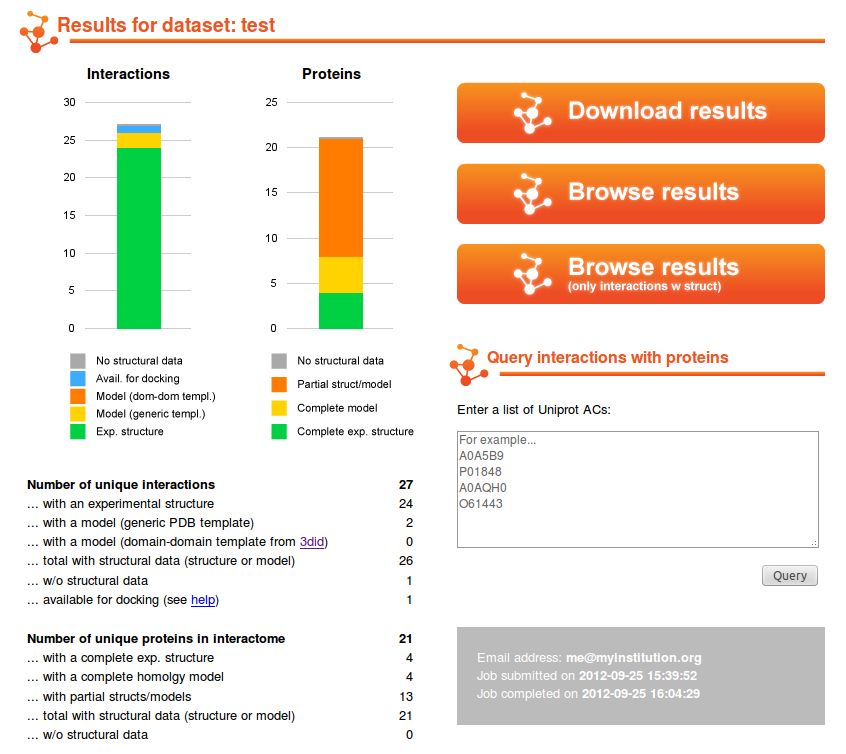
By clicking on the link in your email, you will
be taken to a web page summarizing statistics about your results,
and you will be able to download them, browse them and visualize
a subset of the result involving a list of protein of your
interest. See the following tutorials to learn how to browse the results, query the datasets with a list of proteins of your interest or download the data for offline analysis. |
|
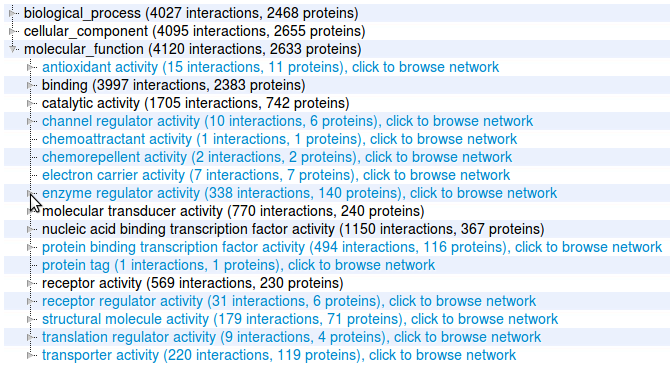
Tutorial: Browse Interactome3D pre-built datasets [top]
1 |
Interactome3D comes with several prebuilt datasets for entire organisms. You can access them through the link of the home page of Interactome3D. Select for instance Mus musculus. |

|
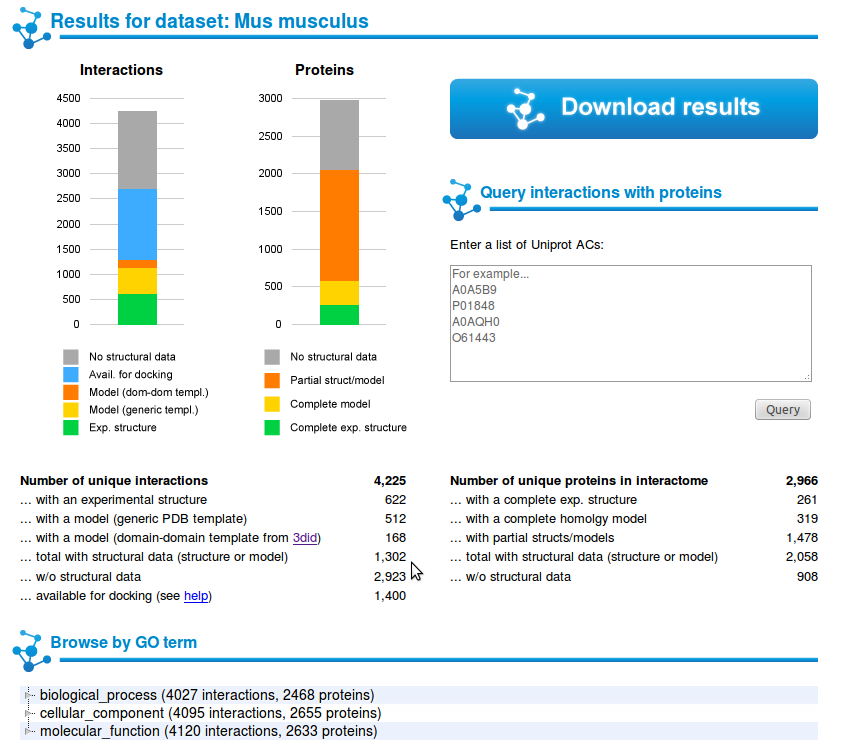
2 |
You will be taken to a page with statistics about your results, and you will be able to download them, browse them, visualize a subset of the result involving a list of proteins of your interest or browse a subset based on GO term annotations. |
|
4 |
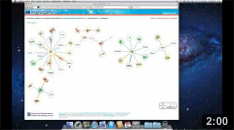
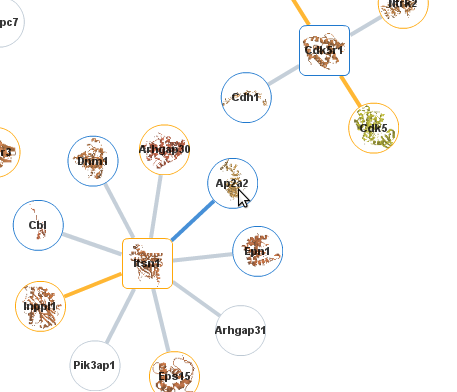
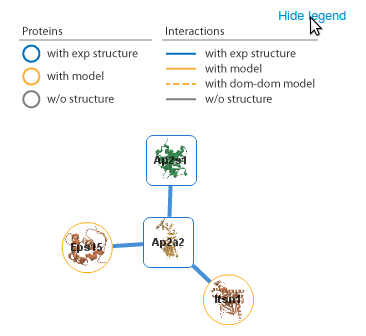
By clicking on one of the links you are taken
to a page that displays interactions involving those proteins.
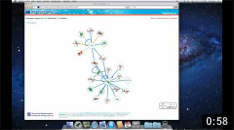
The proteins are represented by circles and the interactions by the
edges that connect them. Proteins and interactions with
structural data are highlighted in the network (see our help page for details). Here we click on the node Ap2A2. |
|
5 |
By clicking on a node you can display details of the structures available for the corresponding proteins, preview them in a window and download them (see our help page for details). |

|
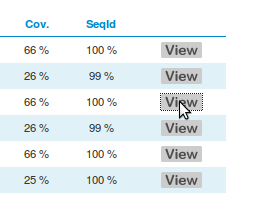
6 |
If you click on one of the View buttons, the corresponding structure will be displayed on the Jmol view at the bottom right corner of the page. |

|
7 |
You can select as many structures and models as you wish and download them. Look at our help page for a description of the file formats. |

|
8 |
You can also click on the edge connecting two
proteins and you will be taken to a page summarizing the structural
data available for this interaction (see our help page for details). Here we clicked on the edge between proteins Ap2A2 and Ap2s1. |

|
9 |
In the bottom right area of the page a box displays the structure of the interaction under analysis. |

|
10 |
In the top right area of the page a box displays the interactome neighborhood of the protein or interaction under analyses (see our help page for details). |
|
11 |
You can go back to the original network by clicking on the Back to full network link at any time. |

|
Tutorial: Query Interactome3D with a list of proteins [top]
1 |
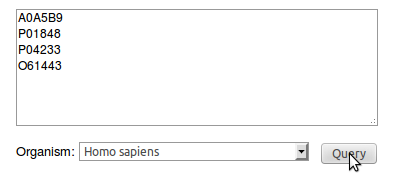
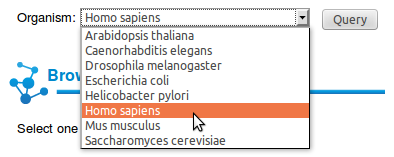
You can query Interactome3D with a list of
proteins of your interest. On the main page you can paste a list of
proteins specified as Uniprot accession codes and choose the
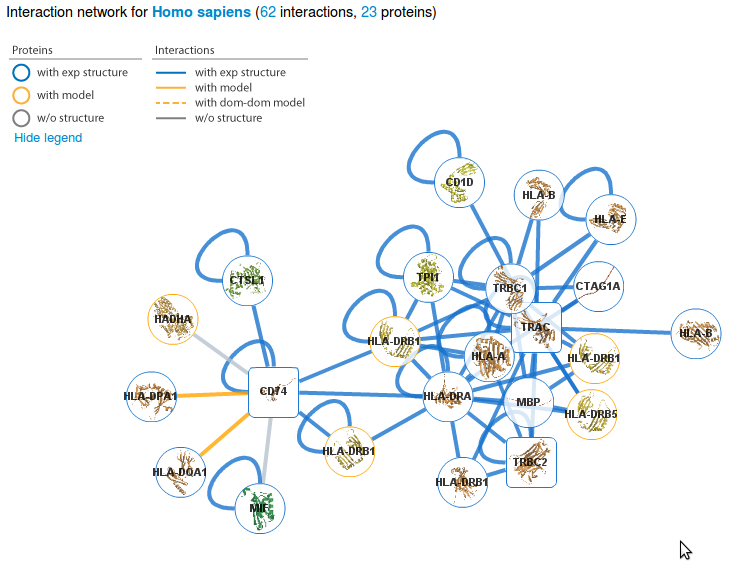
organism you want to look at. Here we select the proteins A0A5B9 P01848 P04233 O61443and the organism Homo sapiens. |
|
2 |
A page will show you the interactions involving your proteins and allow you to see details about proteins and interactions with structural data. Look at the Network help page for details about the network view and to the previous tutorial to learn about the next steps. |
|
3 |
Alternatively, you can query the dataset for an organism directly from its page (See the tutorial Browse Interactome3D pre-built datasets). |
Tutorial: Download Interactome3D data [top]
1 |
The data contained in Interactome3D is also available for download and can be used for offline analyses. |

|
2 |
From the download page you can access the repositories for the different organisms. |

|
3 |
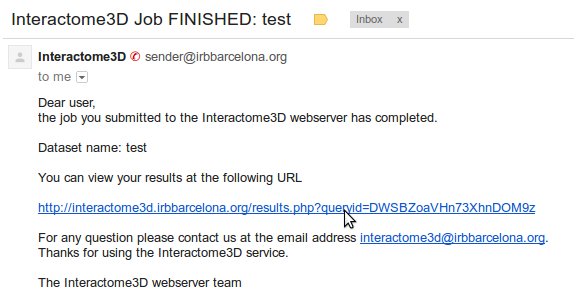
If you submitted your own set of interactions, you will be able to download your results from the summary page, accessible through the link in the confirmation email that you received. |

|
4 |
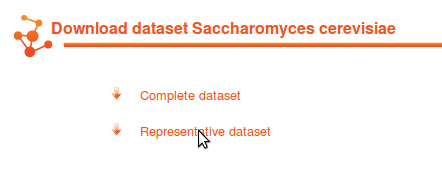
Two datasets are available. One contains the complete set of structures and models for interactions and one contains only a set of representative structures. |
|
5 |
Look at the online documentation to find out how the different datasets are built and the format of the downloadable files. |

|